Genomics
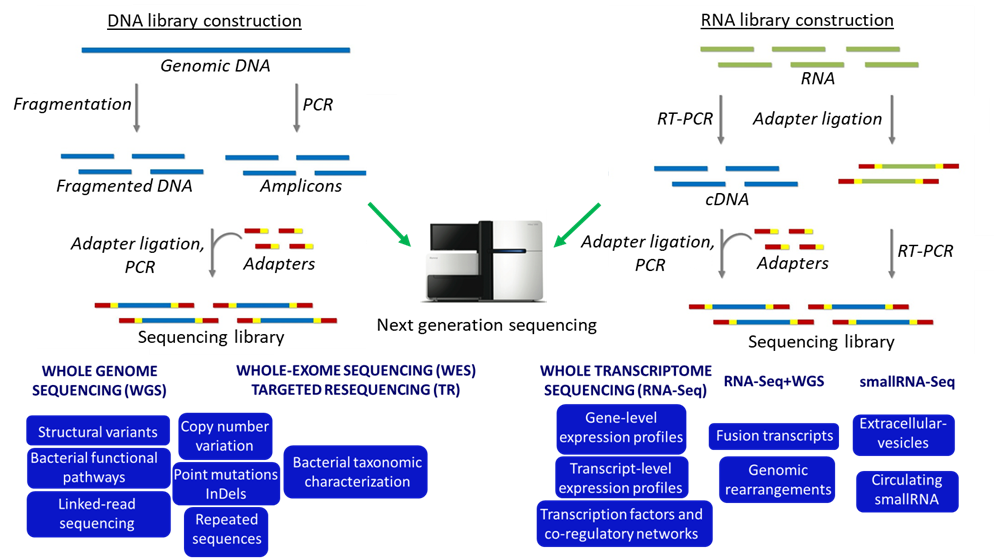
The laboratory of Genomics has a long-standing high-level expertise in the study of the genome by sequencing, mapping and analyzing all genes in a cell, working on a large scale by systematic approaches, the so-called “high-throughput” technologies or next-generation sequencing (NGS).
These NGS-based “-Omics” approaches (Genomics, Transcriptomics, Single-cell analysis, Metagenomics and related Bioinformatics) are applied in order to investigate the molecular mechanisms and interactions involved in health and diseased states.
The Genomic Unit is involved in molecular studies addressing genes at several levels, starting from their base sequence (polymorphism and mutation analysis) and chromosomal re-arrangement (numerical and structural aberration analysis), to, then, expand towards the investigation of their expression, function and reciprocal interactions within the complex network of molecular processes characterizing any biological system (“functional genomics”).
RESEARCH TOPICS
DNA Resequencing combined with capture analysis of target regions was applied to the study of Neurofibromatosis, Noonan Syndrome and Brugada Syndrome. This approach has also been extended to the analysis of the whole exome, where the great majority of disease-related mutations has been located. Single-cell RNA sequencing has been carried out to identify transcriptome signatures in mice with chronic epilepsy caused by a mutation in the Arx gene.
referents: Roberta Bordoni, Eleonora Mangano, Clarissa Consolandi
DNA Resequencing combined with target enrichment was used to study Amyotrophic Lateral Sclerosis (ALS) and Multiple Sclerosis (MS). Whole Genome Sequencing (WGS) of 70 ALS individuals was applied to the systematic analysis of tandem repeats polymorphisms. Resequencing of target regions derived from previous GWAS studies was employed to investigate causal variants in 588 MS patients and 408 healthy controls.
referents: Roberta Bordoni, Eleonora Mangano
Cancer genomics aims to identify new markers for personalized medicine applications. High-throughput technologies, such as NGS and microarrays, allow the comprehensive characterization of single nucleotide variants and chromosomal rearrangements, the expression profiling of both coding and non-coding genes, the identification of particular transcripts, such as fusion transcripts and circular RNAs, till to miRNA expression profiling of circulating extracellular vesicles released by tumor cells.
referents: Ingrid Cifola, Eleonora Mangano, Eva Pinatel, Tania Camboni, Clarissa Consolandi, Francesca Colombo
Genome-wide approach is used for the identification of germline genetic variants associated with the response, both in terms of efficacy and toxicity, to different types of drugs (opioids for cancer pain, immune checkpoint inhibitors, smoking cessation therapies). These studies, part of the so-called precision medicine, aim to stratify patients according to their constitutive characteristics, setting the bases for personalized therapeutic strategies.
referents: Francesca Colombo
Genome plasticity occurs as structural variations like duplications and deletions (1D plasticity), or changes of the positioning of chromosomal domains within the nucleus (3D plasticity). Next-generation sequencing applications allow the study of this plasticity, from the characterization of structural variants and their functional consequences, both in human genome evolution and germline diseases, to the analysis of nuclear architecture during cell differentiation.
referents: Giuliana Giannuzzi and Eva Pinatel
The study of Human Gut Microbiome, by the analysis of the composition and structure by sequencing the 16S gene amplicons and by shotgun sequencing, is applied in several contexts: health and diseased states; in the adaptation studies to different diets and lifestyles: comparison between different populations; in comparison between different eating habits; in volunteers underwent to particular lifestyles; during infections and changes associated with probiotic treatments.
referents: Clarissa Consolandi, Marco Severgnini, Tania Camboni, Camilla Ceccarani
The study of the prokaryotic genome is mainly oriented to identify transcription factor binding sites (ChIP-Seq analyses) and to identify genomic alterations between official strains and newly isolated ones. A customized RNA-seq pipeline has been optimized according to prokaryotic peculiarities and applied to time-course experiments and multiple growth condition comparisons in Helicobacter pylori, Pseudomonas aeruginosa and Streptomyces ambofaciens.
referents: Eva Pinatel, Clarissa Consolandi
STAFF

Roberta Bordoni
Researcher

Tania Camboni
Research Fellow

Camilla Ceccarani
Research Fellow

Ingrid Cifola
Researcher

Francesca Colombo
Researcher

Clarissa Consolandi
Researcher

Gianluca De Bellis
Institute Director Research Executive

Giuliana Giannuzzi
Researcher

Eleonora Mangano
Researcher

Eva Pinatel
Researcher

Marco Severgnini
Researcher