Interactomics
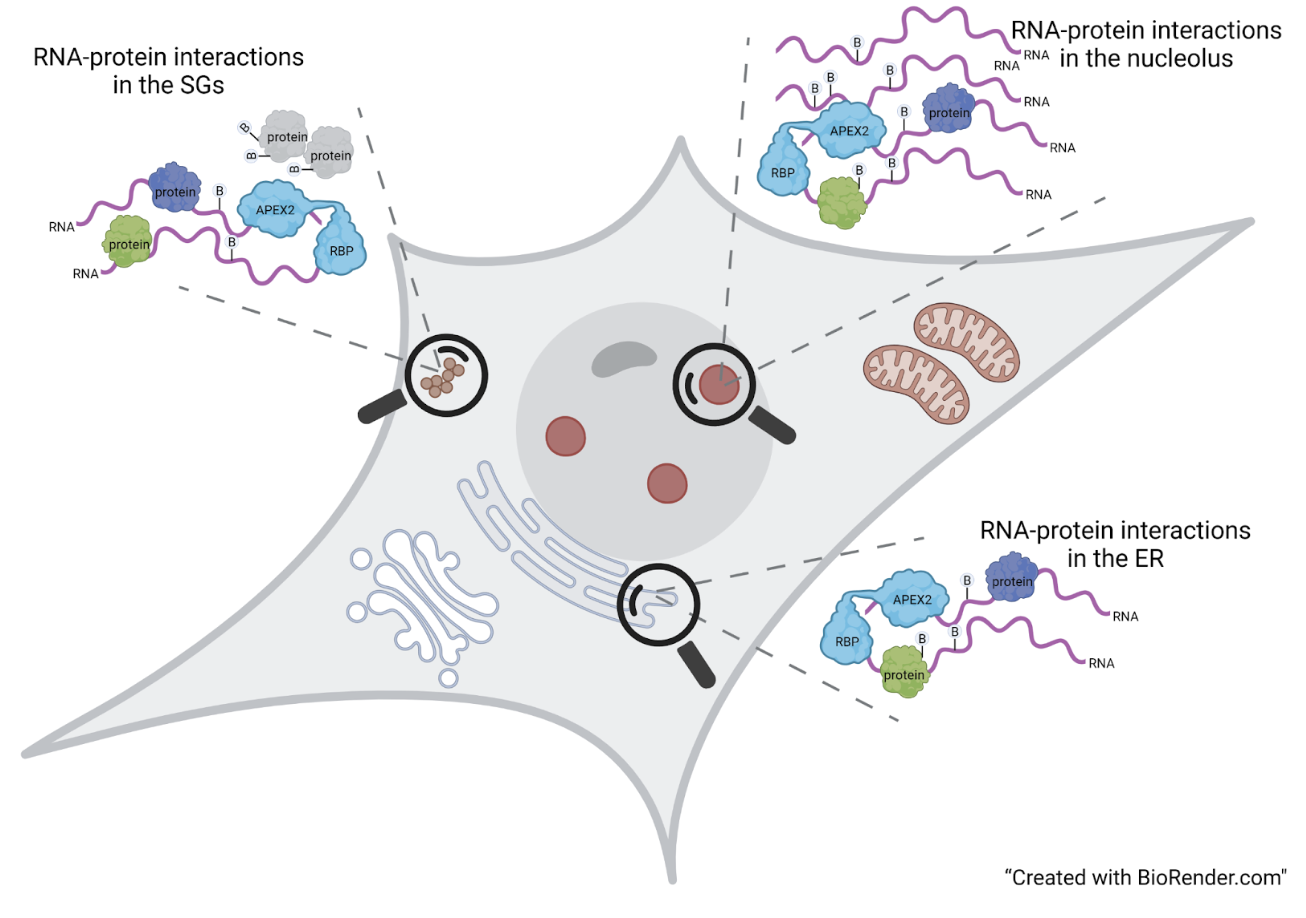
The Interactomics laboratory is concerned with the identification of RNA-protein and protein-protein interactions within mammalian cells. These interactions are essential for maintaining proper cellular homeostasis, and their disruption is linked to numerous human diseases, including neurodegenerative diseases and cancer. Thanks to the development of increasingly sensitive RNA-sequencing and Proteomics approaches, it is now possible to identify most of the interactions occurring in the cell, including transient interactions and between low abundant molecules.The laboratory uses Proximity Ligation technology based on the activity of specific bacterial-derived enzymes that allow it to mark and purify both proteins and RNAs present in close proximity (range < 20 nm) to a protein of interest in living cells.
In particular, through active collaboration with Dr. Francesco Nicassio’s group at the Center for Genomic Science of the Italian Institute of Technology (CGS@SEMM-IIT), the Interactomics group is developing specific approaches for identifying RNAs interacting with various RNA-binding proteins by Nanopore direct-RNA Sequencing.
RESEARCH TOPICS
Through the RNA-Protein Interaction Detection coupled to Mass Spectrometry (RaPID-MS) approach (Ramanathan et al., 2017), we identified proteins that interact with the first and last 1.5 Kb of the viral genome, which are the most structured and containing regulatory regions such as the 5′ and 3′ UTR.
This allowed us to characterize those cellular proteins that interact with and potentially regulate SARS-CoV-2 replication in infected human cells.
We have selected specific RNA-binding proteins (RBPs) that are mutated and whose localization is altered in human neurodegenerative diseases.
Specifically, we want to study which and how the RNAs bound by wild type RBPs compare to their mutated forms in living cells.
We are particularly interested in the study of pseudouridine synthases (PUSs) whose activity converts uridine to pseudouridine. This posttranscriptional modification regulates both the folding of the modified RNA and its recognition and binding by the RBPs of the cell. Through proximity ligation coupled with Nanopore direct-RNA sequencing, we want to identify the substrate RNAs of the different PUSs and further investigate the biological role of pseudouridine in human cells.
STAFF

Roberto Giambruno
Researcher